Dynamical Fluorescence Calcium Imaging in Neuron
We follow calcium concentration in neuron or astrocytes using calcium sensitive fluorescent dyes and propose a denoising procedure and a clustering procedure based on an adaptive statistical comparison of the difference of two time-series to the zero mean vector which follows Baraud, Huet, Laurent (2003) in the independent Gaussian case and Durot, Rozenholc (2006) in a more general case. Using such nonparametric approach avoids modelization of the dynamics of the calcium.
Collaborators Paris Descartes : Christophe Pouzat, Tiffany Lieury
Pre-processing of Fluorescence Calcium Imaging
Keywords : DCE-CT, DCE-MRI, time series, dynamical image, denoising, clustering, Volterra equation of first kind, distribution time transit, Laplace deconvolution, compartmental models, (un-) supervised statistical learning, REMISCAN.
The denoising step builds adaptively around each voxel a 2D or 3D homogenous neighborhood of voxels in the spirit of the Lepskii method, see Rozenholc, Reiss (2012). Three times of the original sequence are shown in their original stage, denoised and by their residuals (difference of original and denoised).
Using the result of the denoising step as input, the clustering procedure runs iteratively from voxels with large neighborhood to voxel with small neighborhood. It stops when all voxels have been clusterized or when a minimal size is reached. As a product, we obtain an automatic clusterization of the tissues with their denoised enhancements. No choice of the number of clusters is needed as input!
See http://www.math-info.univ-paris5.fr/~rozen/RRBC-preprint/last-version.pdf
60 seconds of calcium sensitive fluorescence signal from part of an axon of cerebellar molecular layer interneuron.
R-package : DynClust

The denoised movie obtained with R-package DynClust
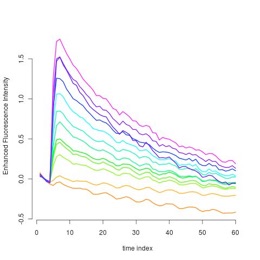
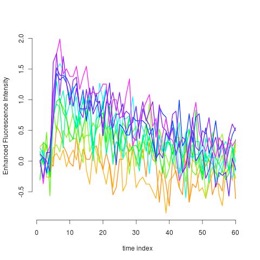
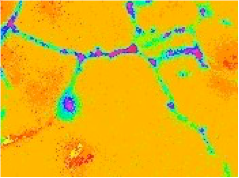
Sample of the original dynamical fluorescence concentrations
in each cluster
Center of the clusters providing denoised representation of the dynamical fluorescence concentrations
Clusters of the dynamical fluorescence concentrations

